Service Overview
Whether you're working with RNA-seq, CRISPR screens, whole-genome sequencing, single-cell data, or any other sequencing technology, we provide expert bioinformatics analysis tailored to your specific research questions. Pay a small deposit to start, then pay based on your data size when you upload.
Example Deliverable: Differential Expression Analysis
Interactive MA plot - click the numbered markers or legend items to learn more
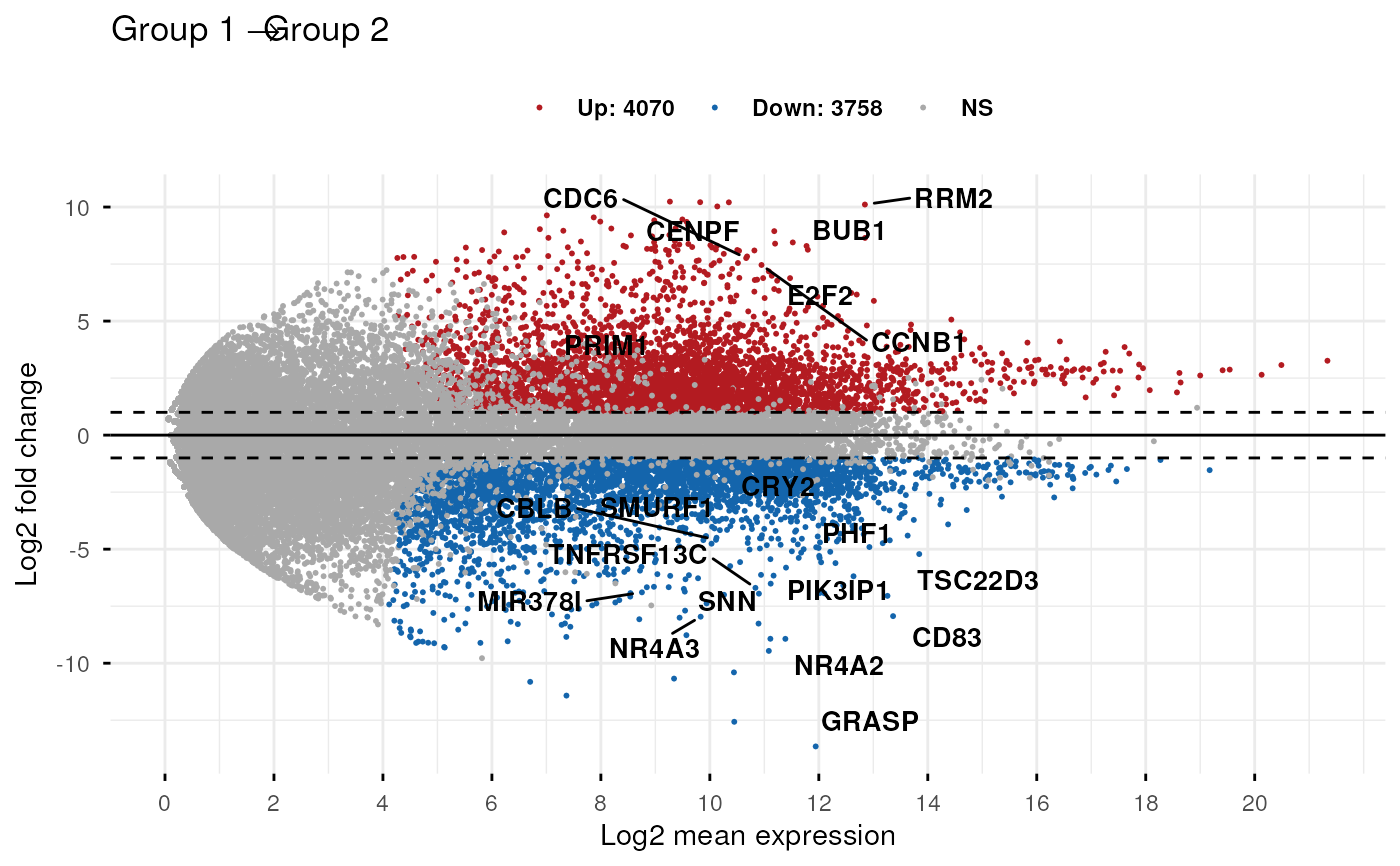
About This Figure
This MA plot visualizes differential gene expression between two experimental conditions. Each dot represents a single gene measured by RNA-sequencing. Click on the numbered markers or legend items above to explore different aspects of the plot.
Click any marker (1-5) or legend item to learn more
Project Types We Support
How It Works
Data Types We Handle
We have experience with virtually any sequencing or omics data type:
What's Included
- Analysis tailored to your specific research questions
- Publication-ready figures (PNG, PDF, SVG)
- Comprehensive written report with interpretation
- Methods section ready for your manuscript
- All processed data files and tables
- Code documentation for reproducibility
- Results walkthrough call (optional)
- Follow-up support after delivery